-Search query
-Search result
Showing 1 - 50 of 63 items for (author: patel & dj)
EMDB-41097: 
cryoEM structure of Smc5/6 5mer
Method: single particle / : Yu Y, Patel DJ

EMDB-41098: 
Smc5/6 8mer
Method: single particle / : Yu Y, Patel DJ

EMDB-26140: 
ATP and DNA bound SMC5/6 core complex
Method: single particle / : Yu Y, Patel DJ

EMDB-24794: 
GLP-1 receptor bound with Pfizer small molecule agonist
Method: single particle / : Liu Y, Dias JM, Han S

EMDB-24292: 
Cryo-EM structure of DNMT5 in apo state
Method: single particle / : Wang J, Patel DJ

EMDB-24294: 
Cryo-EM structure of DNMT5 binary complex with hemimethylated DNA
Method: single particle / : Wang J, Patel DJ

EMDB-24295: 
cryo-EM structure of DNMT5 quaternary complex with hemimethylated DNA, AMP-PNP and SAH
Method: single particle / : Wang J, Patel DJ

EMDB-25577: 
Cryo-EM structure of DNMT5 pseudo-ternary complex solved by incubation with hemimethylated DNA and SAM
Method: single particle / : Wang J, Patel DJ

EMDB-25164: 
Cryo-EM structure of AdnA-AdnB(W325A) in complex with DNA and AMPPNP
Method: single particle / : Wang J, Warren GM, Shuman S, Patel DJ

EMDB-23517: 
Nse5-6 complex
Method: single particle / : Yu Y, Li SB

EMDB-23345: 
Cryo-EM structure of Mycobacterium smegmatis Lhr helicase C-terminal domain
Method: single particle / : Wang J, Warren GM

EMDB-23244: 
Structure of human SHLD2-SHLD3-REV7-TRIP13(E253Q) complex
Method: single particle / : Xie W, Patel DJ

EMDB-30767: 
Cryo-EM structure of LshCas13a-crRNA-anti-tag RNA complex
Method: single particle / : Wang B, Zhang T

EMDB-30453: 
NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP
Method: single particle / : Li W, Tian W, Yuan G, Deng P, Gozani O, Patel D, Wang Z

EMDB-30455: 
NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode)
Method: single particle / : Li W, Tian W, Yuan G, Deng P, Gozani O, Patel D, Wang Z

EMDB-30456: 
NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode)
Method: single particle / : Li W, Tian W, Yuan G, Deng P, Gozani O, Patel D, Wang Z

EMDB-30457: 
Native NSD3 bound to 187-bp nucleosome
Method: single particle / : Li W, Tian W, Yuan G, Deng P, Gozani O, Patel D, Wang Z

EMDB-21366: 
Cryo-EM structure of AcrVIA1-Cas13(crRNA) complex
Method: single particle / : Jia N, Meeske AJ

EMDB-21367: 
Cryo-EM structure of Cas13(crRNA)
Method: single particle / : Jia N, Meeske AJ

EMDB-21126: 
Cryo-EM structure of Cascade-TniQ binary complex
Method: single particle / : Jia N, Patel DJ

EMDB-21146: 
Cryo-EM structure of Cascade-TniQ-dsDNA ternary complex
Method: single particle / : Jia N, Patel DJ

EMDB-20440: 
Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP
Method: single particle / : Jia N, Unciuleac M, Shuman S, Patel DJ

EMDB-20446: 
Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP and DNA
Method: single particle / : Jia N, Unciuleac M, Shuman S, Patel DJ

EMDB-20447: 
Cryo-EM structure of AdnAB-AMPPNP-DNA complex
Method: single particle / : Jia N, Unciuleac M, Shuman S, Patel DJ

EMDB-0640: 
Cryo-EM structure of Csm-crRNA-target RNA ternary complex in complex with AMPPNP in type III-A CRISPR-Cas system
Method: single particle / : Jia N, Patel DJ

EMDB-0641: 
Cryo-EM structure of Csm-crRNA-target RNA ternary complex in complex with cA4 in type III-A CRISPR-Cas system
Method: single particle / : Jia N, Patel DJ

EMDB-0642: 
Cryo-EM structure of Csm-crRNA-target RNA ternary bigger complex in complex with cA4 in type III-A CRISPR-Cas system
Method: single particle / : Jia N, Patel DJ
EMDB-9175: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate 224-13
Method: single particle / : Nogal B, Ward AB
EMDB-9176: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RCn
Method: single particle / : Nogal B, Ward AB
EMDB-9177: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RFr16
Method: single particle / : Nogal B, Ward AB

EMDB-9178: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate ROw16
Method: single particle / : Nogal B, Ward AB

EMDB-9179: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RQq16
Method: single particle / : Nogal B, Ward AB

EMDB-9180: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RRk16
Method: single particle / : Nogal B, Ward AB
EMDB-9181: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RVh16
Method: single particle / : Nogal B, Ward AB
EMDB-9182: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RTh16
Method: single particle / : Nogal B, Ward AB
EMDB-9183: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RWh16
Method: single particle / : Nogal B, Ward AB
EMDB-9184: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RWr16
Method: single particle / : Nogal B, Ward AB
EMDB-9185: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate RYm16
Method: single particle / : Nogal B, Ward AB
EMDB-9186: 
Negative Stain EM map of polyclonal serum in complex with BG505 SOSIP.664 from non-human primate ROw16
Method: single particle / : Nogal B, Ward AB
EMDB-0569: 
HIV-1 vaccine elicited polyclonal 5N6 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB
EMDB-0570: 
HIV-1 vaccine elicited polyclonal 99-13 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0571: 
HIV-1 vaccine elicited polyclonal 145-11 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB
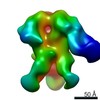
EMDB-0572: 
HIV-1 vaccine elicited polyclonal BM57 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0573: 
HIV-1 vaccine elicited polyclonal BO77 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0574: 
HIV-1 vaccine elicited polyclonal BO77 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB
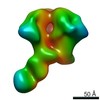
EMDB-0575: 
HIV-1 vaccine elicited polyclonal RAa14 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0576: 
HIV-1 vaccine elicited polyclonal RAv16 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0577: 
HIV-1 vaccine elicited polyclonal RAv16 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB
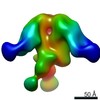
EMDB-0578: 
HIV-1 vaccine elicited polyclonal REj15 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB

EMDB-0579: 
HIV-1 vaccine elicited polyclonal REv16 Fab in complex with the HIV Env trimer BG505 SOSIPv5.2
Method: single particle / : Nogal B, Ward AB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
